n <- 1000
p <- 3
X1 <- matrix(rnorm(n * p), ncol = p)
X2 <- X1
X2[, 1] <- X2[, 1] ^ 2
beta <- rep(1, p)
y_logit <- X2 %*% beta
p <- plogis(y_logit)
outcome <- rbinom(n * p, 1, p)Assessing linearity in logistic regression
Background
https://chatgpt.com/share/6851c5ad-9c08-8013-ad26-d03c0de42079
Simulate some data
Fit a model
fm1 <- glm(outcome ~ X1, family = binomial())
fm2 <- glm(outcome ~ X2, family = binomial())Partial residual plots
library(faraway)
prplot(fm1, 1)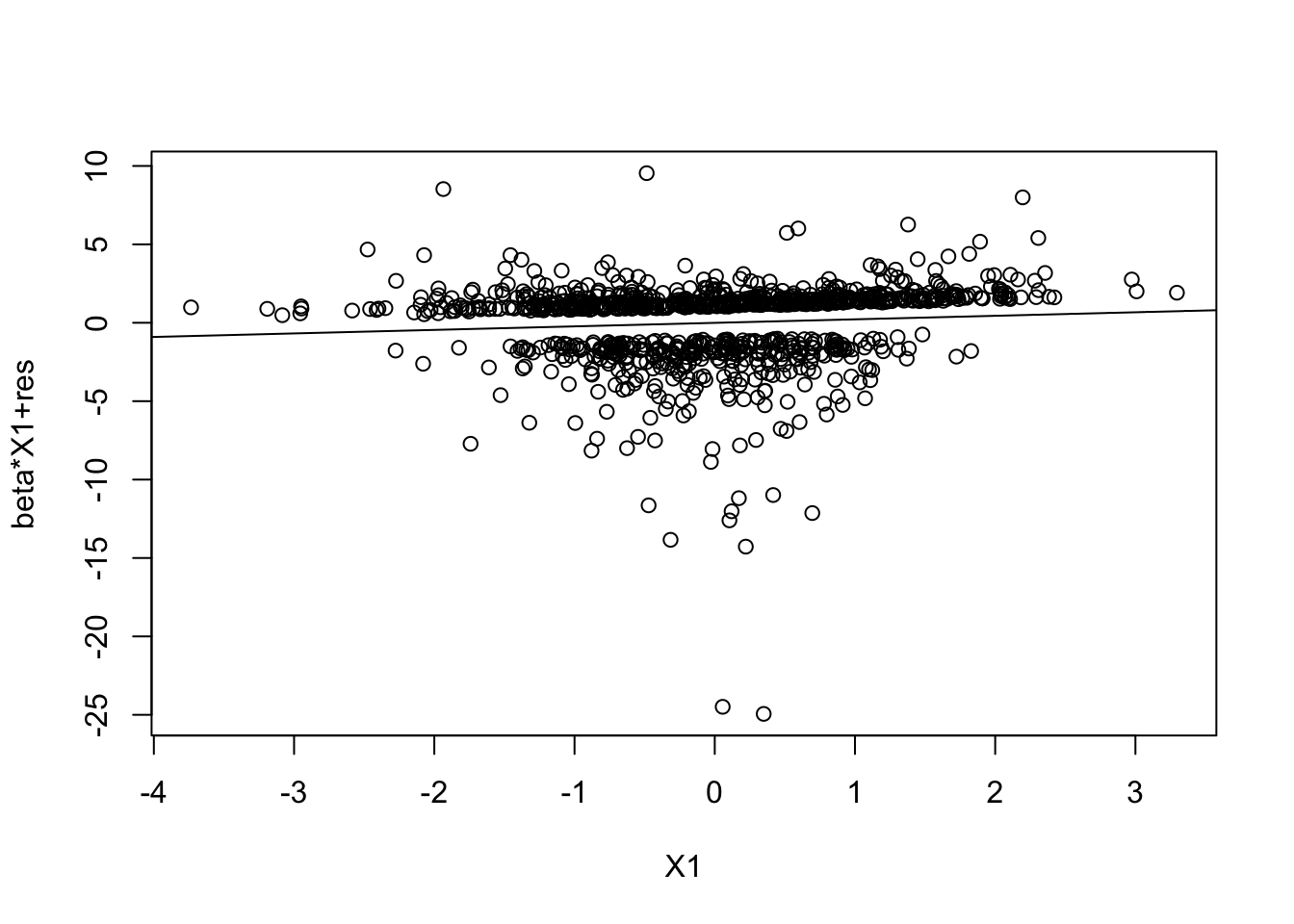
prplotfunction (g, i)
{
xl <- attributes(g$terms)$term.labels[i]
yl <- paste("beta*", xl, "+res", sep = "")
x <- model.matrix(g)[, i + 1]
plot(x, g$coeff[i + 1] * x + g$res, xlab = xl, ylab = yl)
abline(0, g$coeff[i + 1])
invisible()
}
<bytecode: 0x10fbf0978>
<environment: namespace:faraway>library('tidyverse')── Attaching core tidyverse packages ──────────────────────── tidyverse 2.0.0 ──
✔ dplyr 1.1.4 ✔ readr 2.1.5
✔ forcats 1.0.0 ✔ stringr 1.5.1
✔ ggplot2 3.5.1 ✔ tibble 3.2.1
✔ lubridate 1.9.4 ✔ tidyr 1.3.1
✔ purrr 1.0.2
── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
✖ dplyr::filter() masks stats::filter()
✖ dplyr::lag() masks stats::lag()
ℹ Use the conflicted package (<http://conflicted.r-lib.org/>) to force all conflicts to become errorsprplot2 <- function (g, i)
{
xl <- attributes(g$terms)$term.labels[i]
yl <- paste("beta*", xl, "+res", sep = "")
x <- model.matrix(g)[, i + 1]
y <- g$coeff[i + 1] * x + g$res
tibble(x = x, y = y) |>
ggplot(aes(x, y)) + geom_point() + geom_smooth()
}
prplot2(fm1, 1)`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'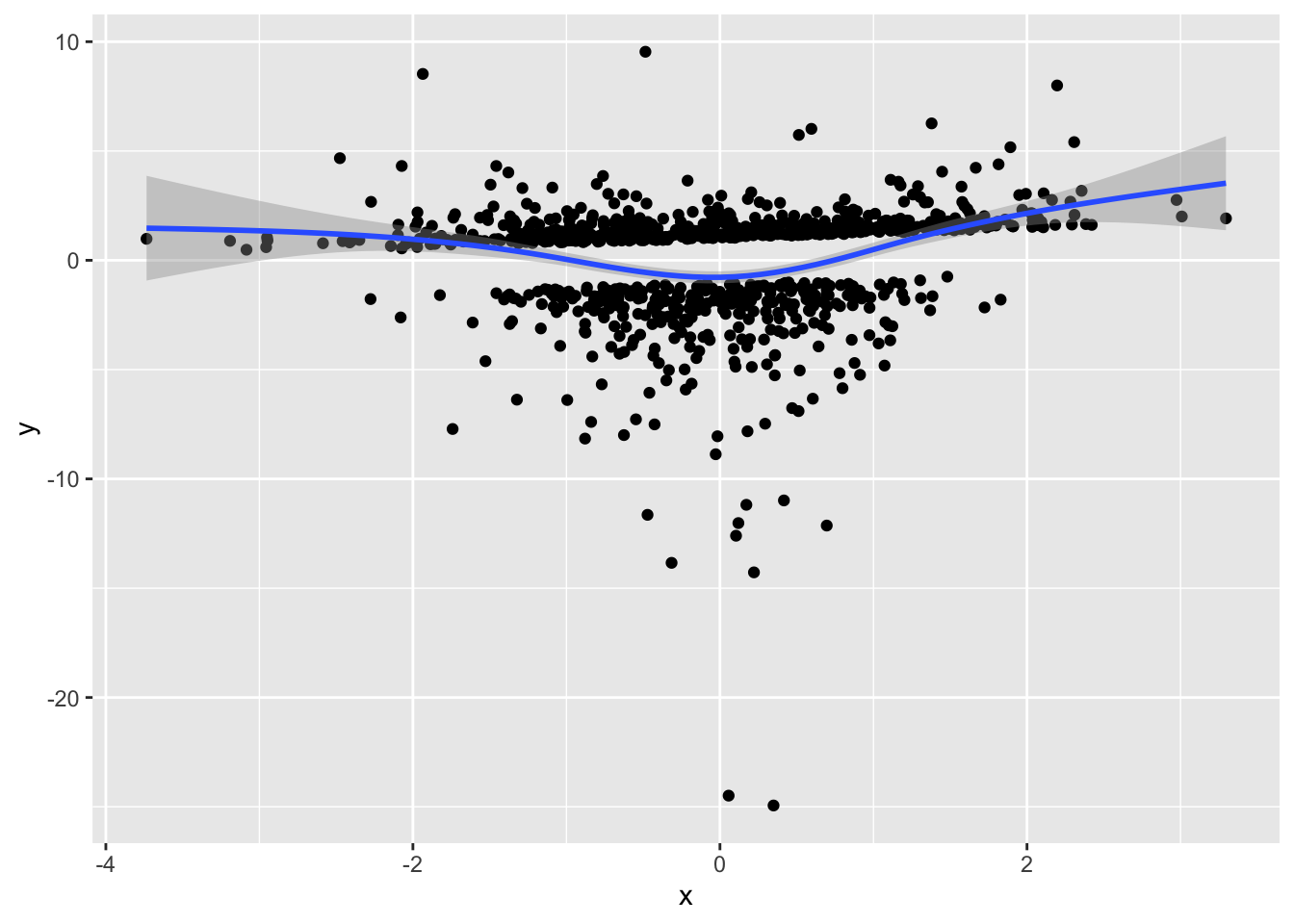
prplot2(fm2, 1)`geom_smooth()` using method = 'gam' and formula = 'y ~ s(x, bs = "cs")'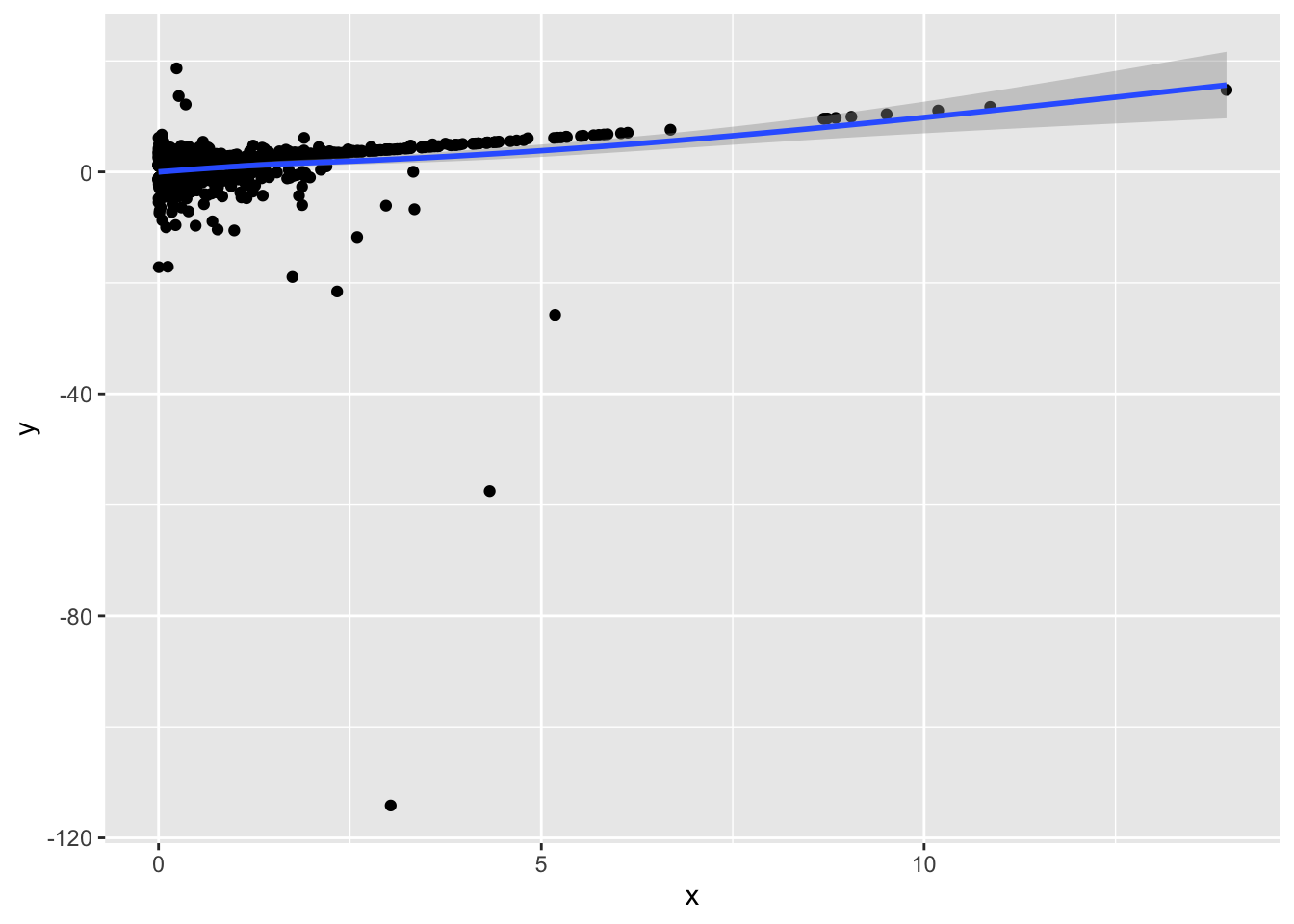
Binned plot
library(arm)Loading required package: MASS
Attaching package: 'MASS'The following object is masked from 'package:dplyr':
selectLoading required package: Matrix
Attaching package: 'Matrix'The following objects are masked from 'package:tidyr':
expand, pack, unpackLoading required package: lme4
arm (Version 1.14-4, built: 2024-4-1)Working directory is /Users/seb/Documents/Projects/00_websites/sbloggel/posts/2025-07-30-assessing-linearity-in-logistic-regression
Attaching package: 'arm'The following objects are masked from 'package:faraway':
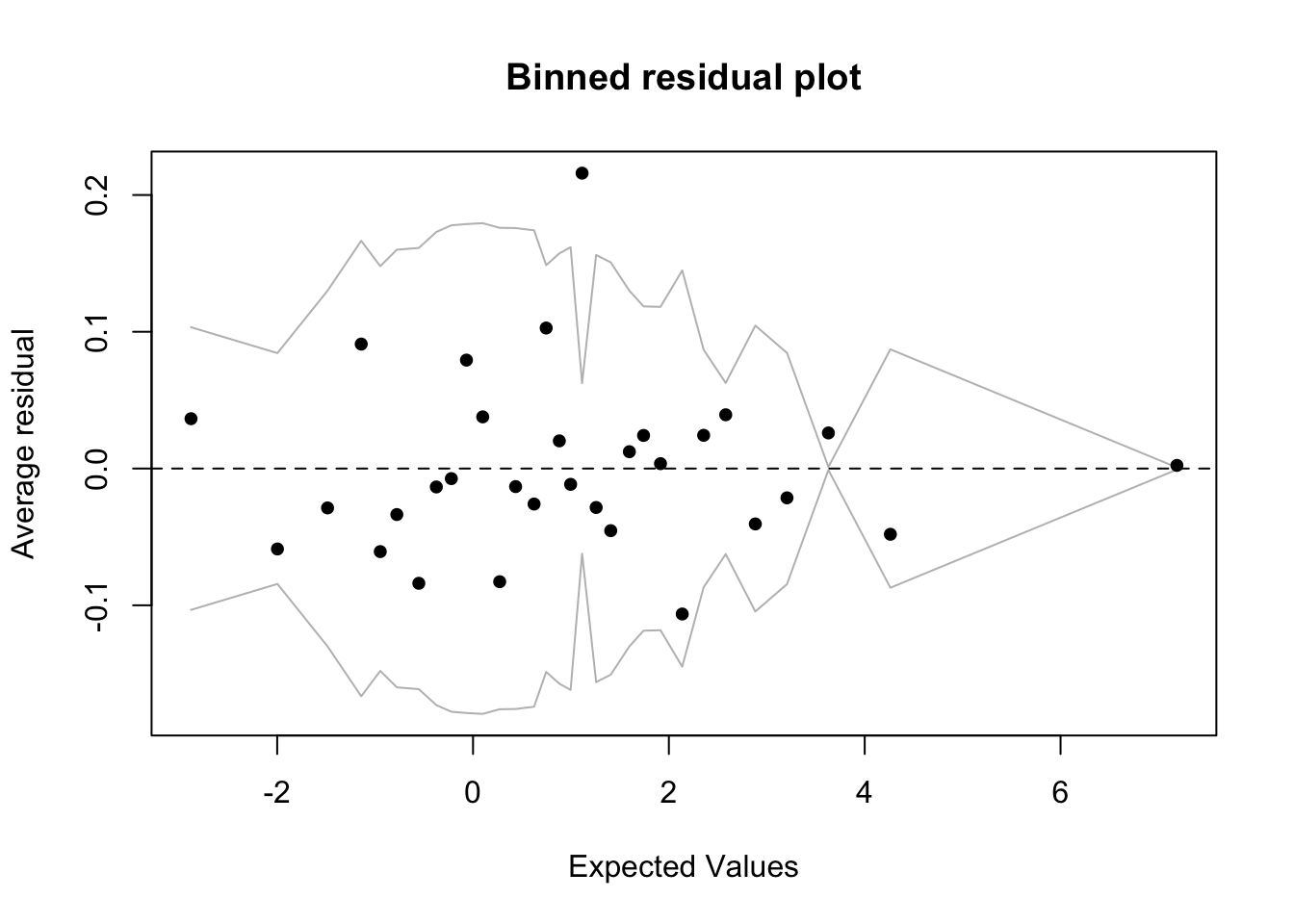
fround, logit, pfroundbinnedplot(predict(fm1), residuals(fm1, type = "response"))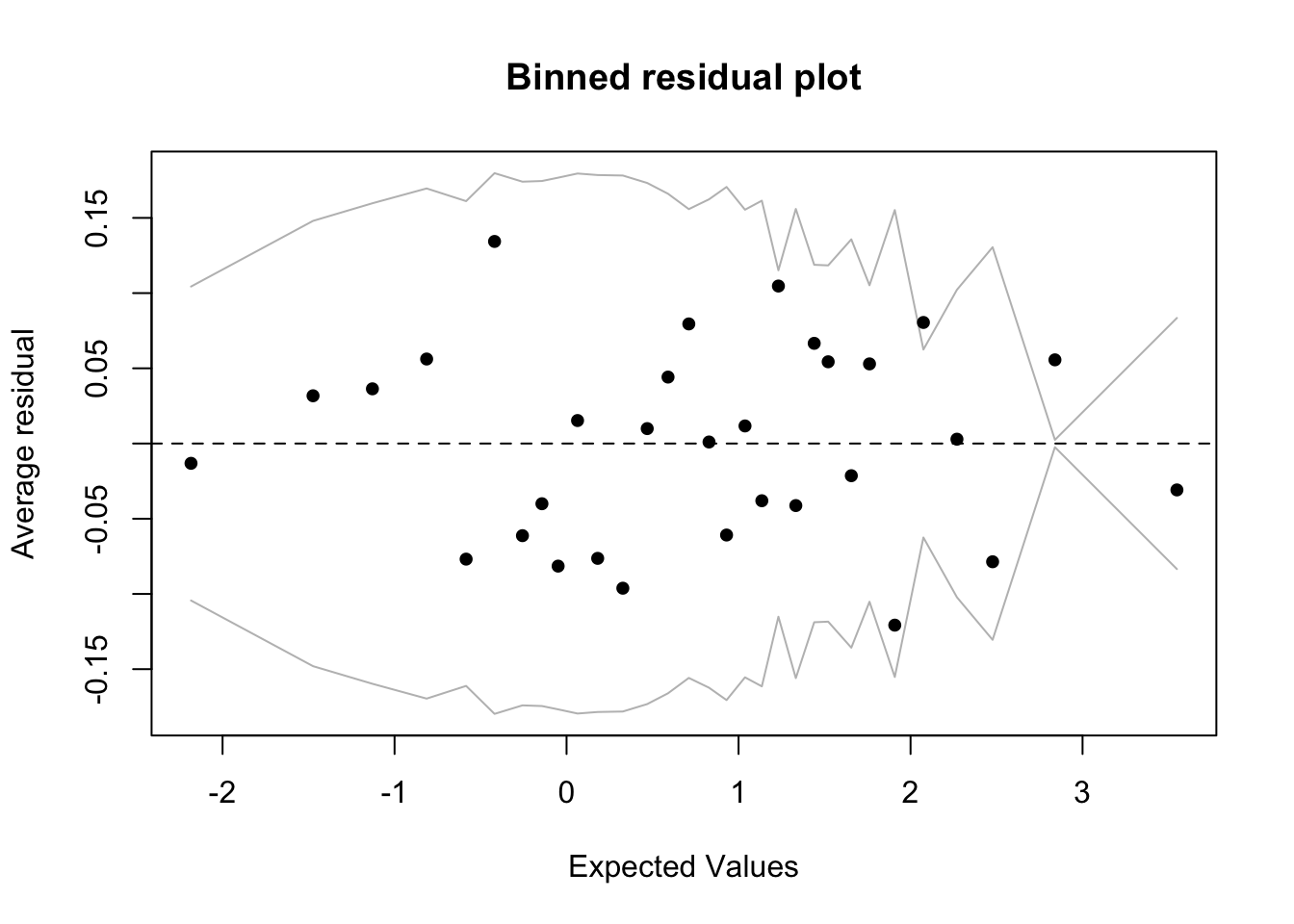
binnedplot(predict(fm2), residuals(fm2, type = "response"))