library('tidyverse')
library('faraway')
set.seed(1)Model diagnostics for logistic regression
n <- 100
dat <- tibble(x1 = rnorm(n),
x2 = rnorm(n),
x3 = rnorm(n),)
X <- model.matrix(~ ., data = dat)
beta <- rnorm(ncol(X))
dat <- dat |>
mutate(mu = X %*% beta,
p = plogis(mu),
result = rbinom(n, 1, p))
fitted_model <- glm(result ~ x1 + x2 + x3, data = dat, family = binomial())
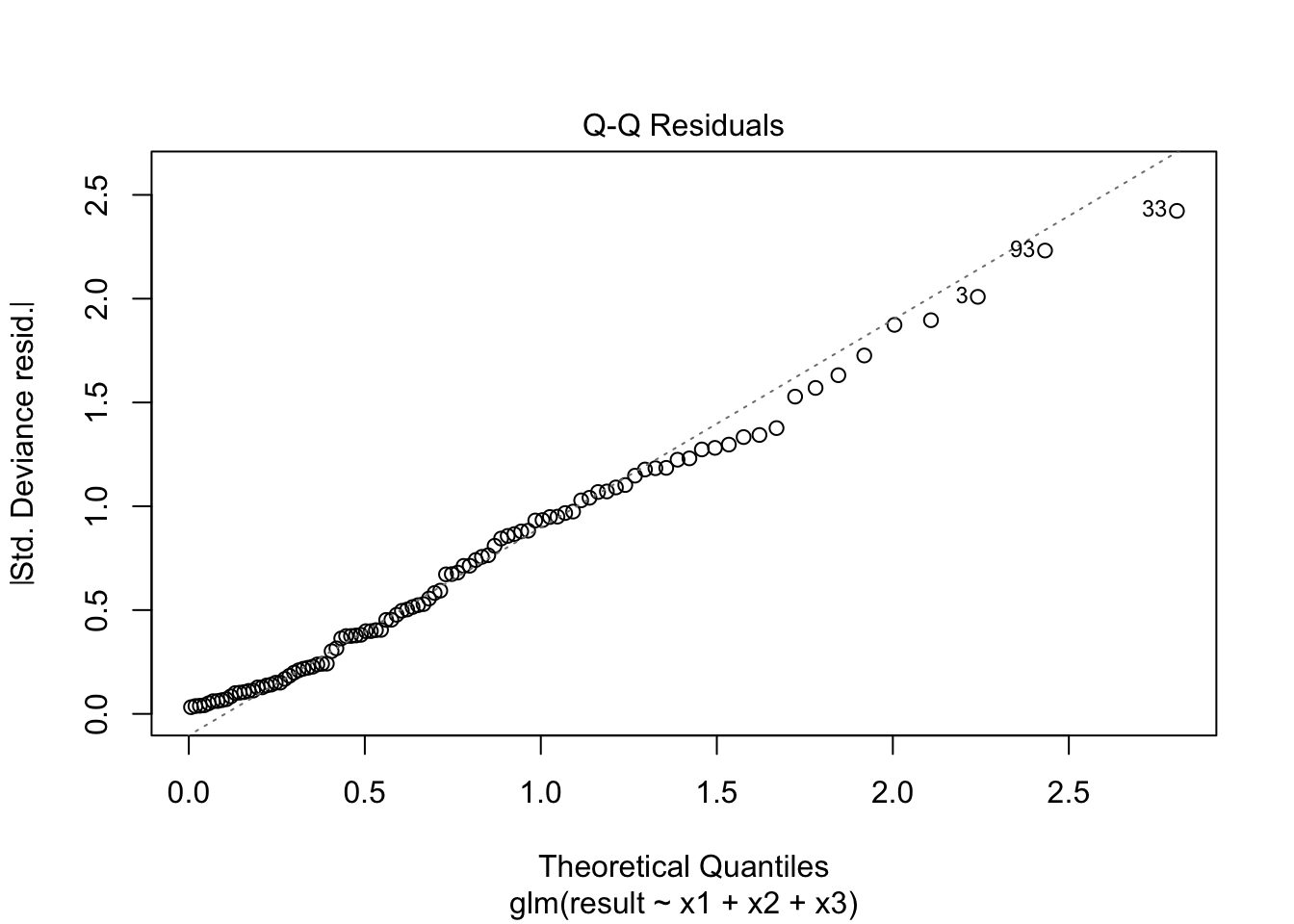
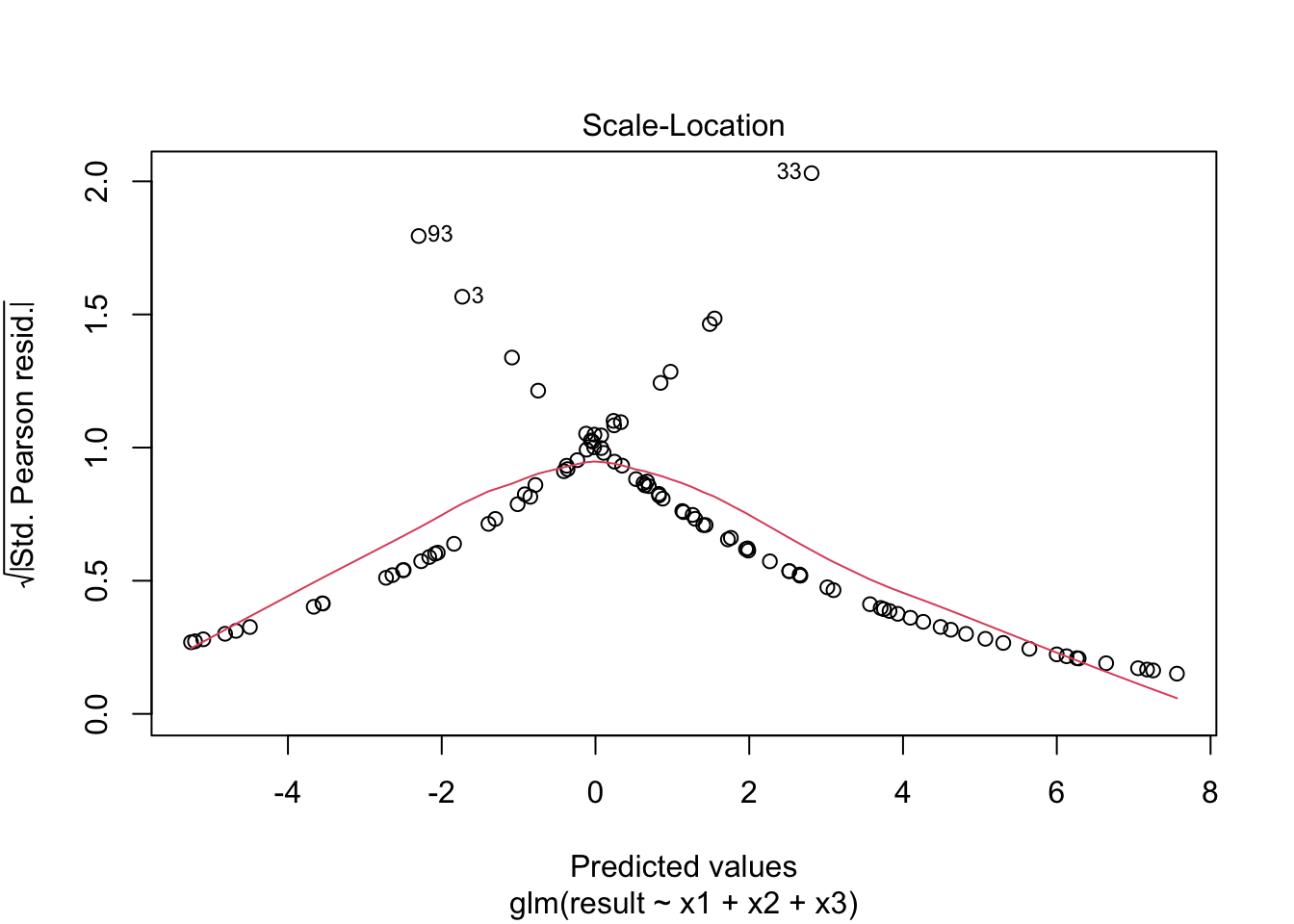
plot(fitted_model)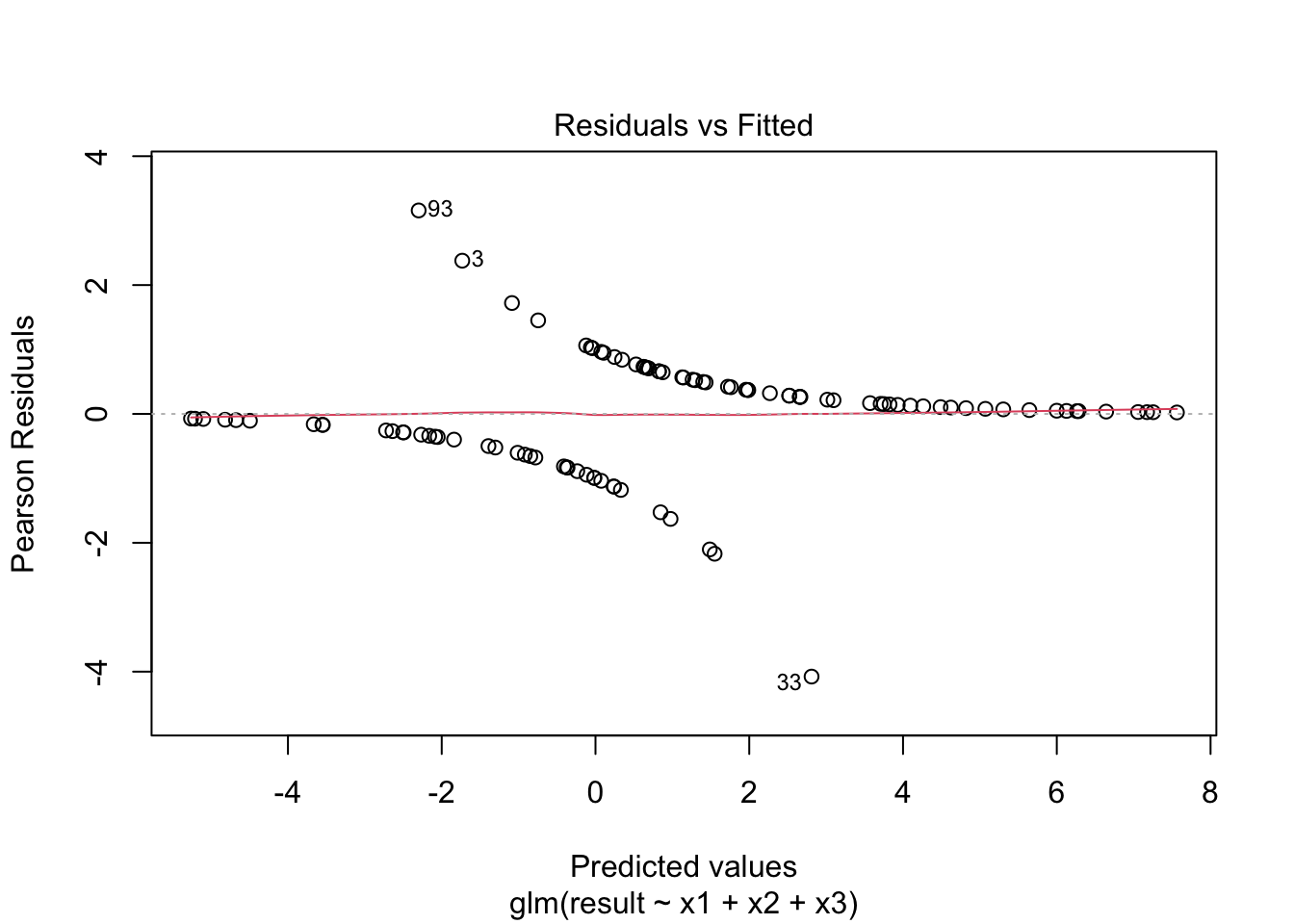
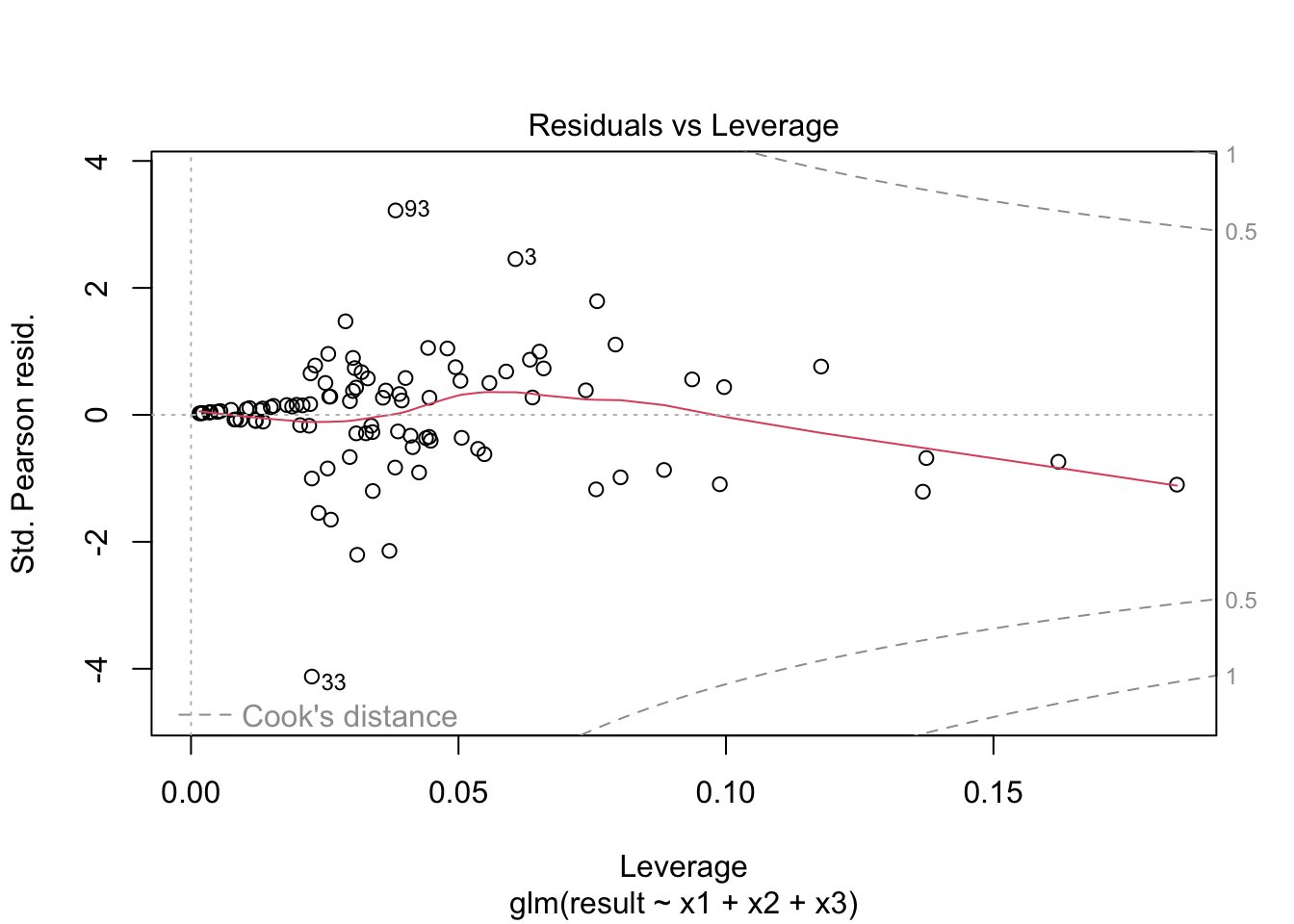
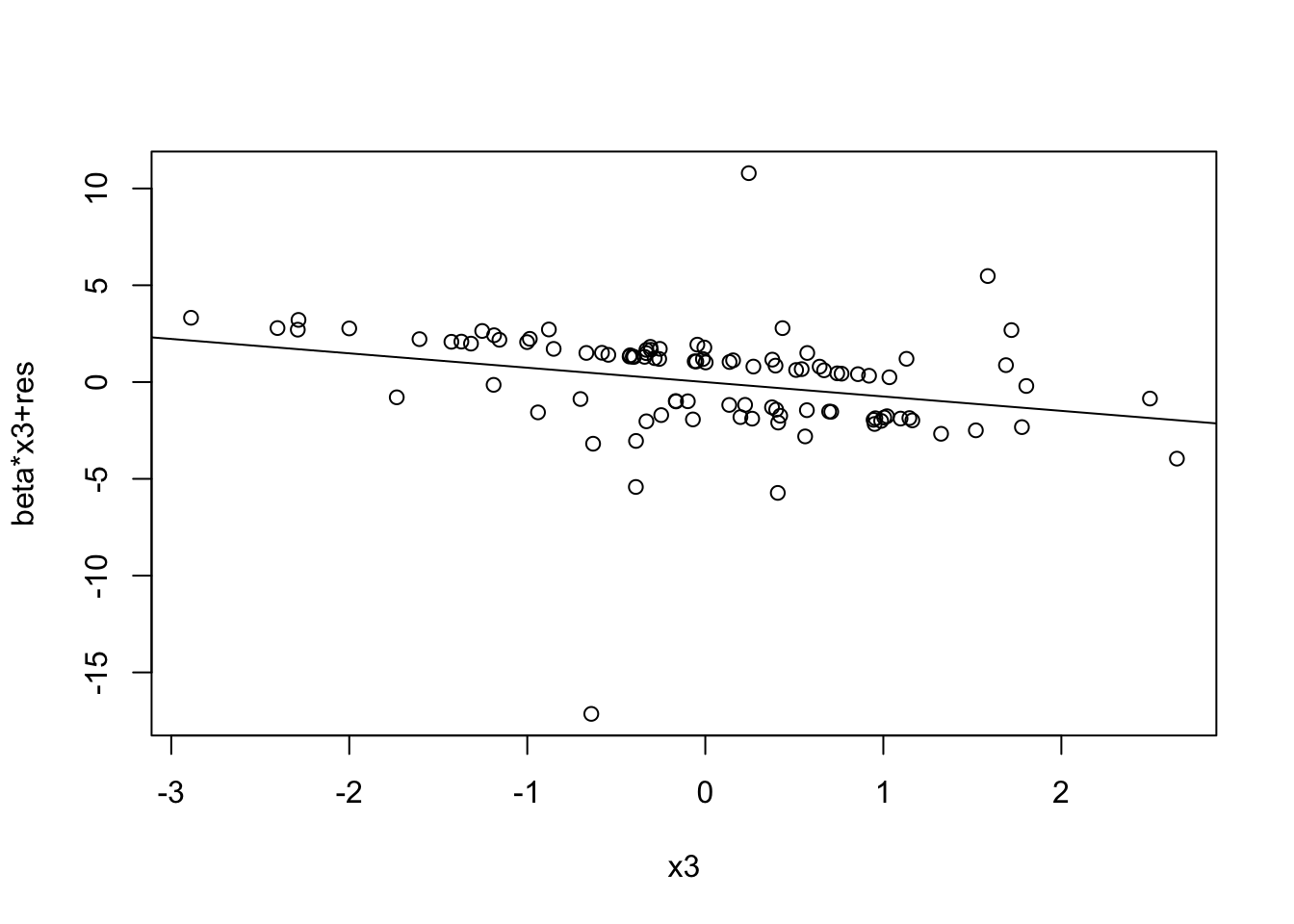
prplot(fitted_model, 1)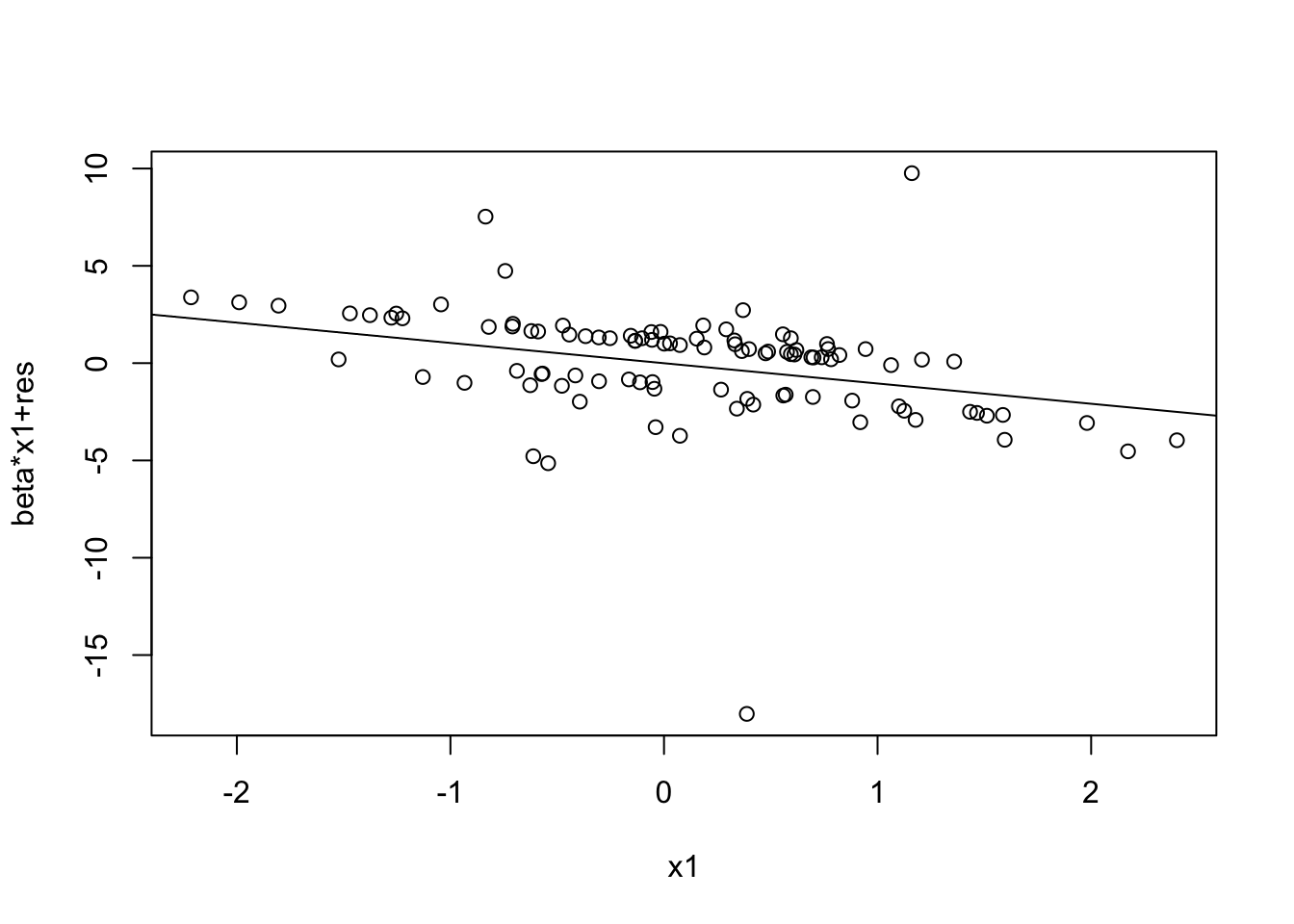
prplot(fitted_model, 2)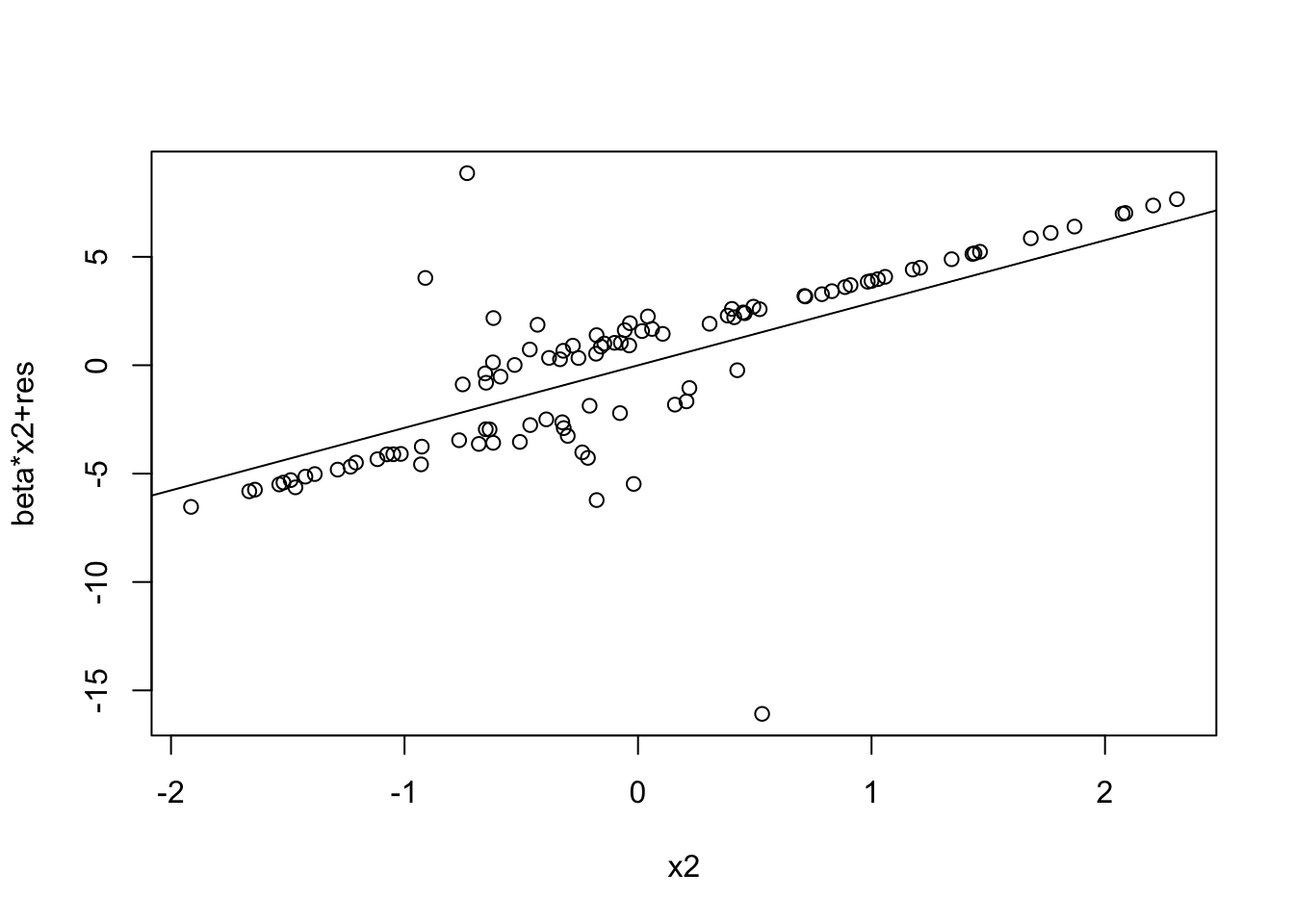
prplot(fitted_model, 3)