# size of sample:
n <- 20
m <- 5
# model parameters:
beta_0 <- 0
beta_1 <- 1
sigma_0 <- 2
sigma_1 <- 1
sigma_2 <- 5
# construct data:
patients <- tibble(patient_id = 1:n,
patient_re = rnorm(n, 0, sigma_1),
treatment_re = rnorm(n, 0, sigma_2))
dat <- expand_grid(patients,
replicate = 1:m,
timepoint = c('pre', 'post'))
dat <- dat |>
mutate(y = beta_0 +
patient_re +
ifelse(timepoint == 'post', treatment_re + beta_1, 0) +
rnorm(n * m * 2, 0, sigma_0))
# make timepoint a factor to get order in plots right:
dat <- dat |>
mutate(timepoint = factor(timepoint, levels = c('pre', 'post')))Specifying mixed effects in lme4
Description of trial
Underlying probability model
Notation
Parameters of the model to be estimated
Model equations
- Measurement pre intervention:
- Measurement post-intervention:
Simulate data
Tabular characteristics
dat |>
group_by(timepoint) |>
summarize(mean = mean(y)) |>
kable()| timepoint | mean |
|---|---|
| pre | 0.1868025 |
| post | 1.1930561 |
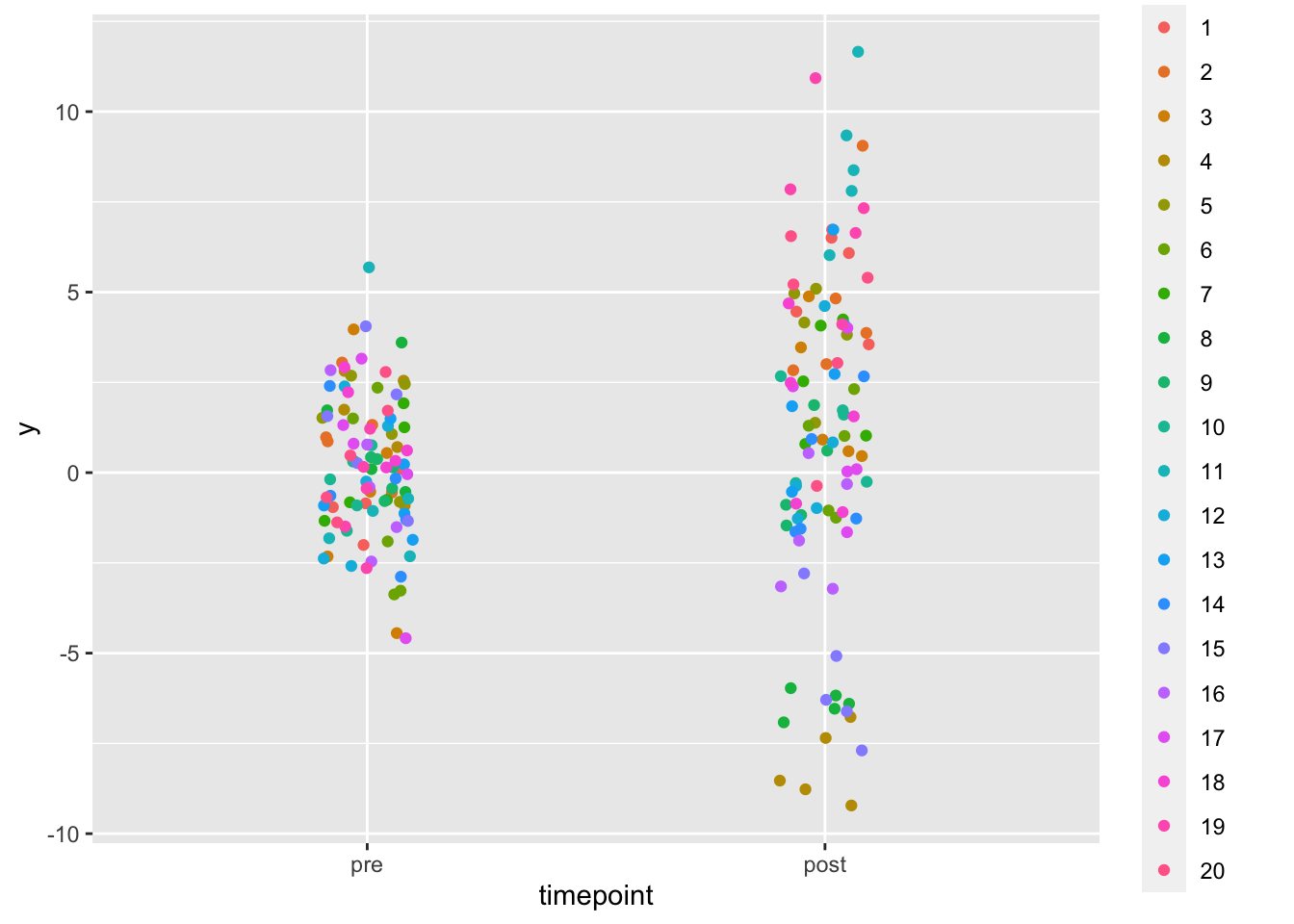
Visualize data
if(n < 1e2) {
dat |>
ggplot(aes(timepoint, y, col = factor(patient_id))) +
geom_jitter(width = 0.1, height = 0) + labs(color = 'Patient ID')
}
Fit model
Linear model
# linear model to get beta_1 and beta_2
(fm_lm <- lm(y ~ timepoint, data = dat)) |> summary()
Call:
lm(formula = y ~ timepoint, data = dat)
Residuals:
Min 1Q Median 3Q Max
-10.4127 -1.5931 0.0072 2.0727 10.4653
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 0.1868 0.3489 0.535 0.5930
timepointpost 1.0063 0.4934 2.039 0.0427 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 3.489 on 198 degrees of freedom
Multiple R-squared: 0.02057, Adjusted R-squared: 0.01563
F-statistic: 4.159 on 1 and 198 DF, p-value: 0.04274Mixed effects model with correlation between (Intercept) and timepointpost
(fm_lmer <- lmer(y ~ timepoint + (timepoint | patient_id), data = dat))boundary (singular) fit: see help('isSingular')Linear mixed model fit by REML ['lmerMod']
Formula: y ~ timepoint + (timepoint | patient_id)
Data: dat
REML criterion at convergence: 888.5308
Random effects:
Groups Name Std.Dev. Corr
patient_id (Intercept) 0.3268
timepointpost 4.5307 -1.00
Residual 1.9086
Number of obs: 200, groups: patient_id, 20
Fixed Effects:
(Intercept) timepointpost
0.1868 1.0063
optimizer (nloptwrap) convergence code: 0 (OK) ; 0 optimizer warnings; 1 lme4 warnings Mixed effects model without correlation between (Intercept) and timepointpost
- The specifications I tried seem almost right, but not quite:
(fm_lmer <- lmer(y ~ timepoint + (timepoint - 1 || patient_id), data = dat))Linear mixed model fit by REML ['lmerMod']
Formula: y ~ timepoint + ((0 + timepoint | patient_id))
Data: dat
REML criterion at convergence: 888.5308
Random effects:
Groups Name Std.Dev. Corr
patient_id timepointpre 0.3268
timepointpost 4.2038 -1.00
Residual 1.9086
Number of obs: 200, groups: patient_id, 20
Fixed Effects:
(Intercept) timepointpost
0.1868 1.0063 (fm_lmer <- lmer(y ~ timepoint + (timepoint - 1 | patient_id), data = dat))Linear mixed model fit by REML ['lmerMod']
Formula: y ~ timepoint + (timepoint - 1 | patient_id)
Data: dat
REML criterion at convergence: 888.5308
Random effects:
Groups Name Std.Dev. Corr
patient_id timepointpre 0.3268
timepointpost 4.2038 -1.00
Residual 1.9086
Number of obs: 200, groups: patient_id, 20
Fixed Effects:
(Intercept) timepointpost
0.1868 1.0063 (fm_lmer <- lmer(y ~ timepoint + (1 | patient_id) + (timepoint - 1 | patient_id), data = dat))boundary (singular) fit: see help('isSingular')Linear mixed model fit by REML ['lmerMod']
Formula: y ~ timepoint + (1 | patient_id) + (timepoint - 1 | patient_id)
Data: dat
REML criterion at convergence: 888.5308
Random effects:
Groups Name Std.Dev. Corr
patient_id (Intercept) 0.0000
patient_id.1 timepointpre 0.3268
timepointpost 4.2039 -1.00
Residual 1.9086
Number of obs: 200, groups: patient_id, 20
Fixed Effects:
(Intercept) timepointpost
0.1868 1.0063
optimizer (nloptwrap) convergence code: 0 (OK) ; 0 optimizer warnings; 1 lme4 warnings (fm_lmer <- lmer(y ~ timepoint + (1 | patient_id) + (timepoint + 0 | patient_id), data = dat))boundary (singular) fit: see help('isSingular')Linear mixed model fit by REML ['lmerMod']
Formula: y ~ timepoint + (1 | patient_id) + (timepoint + 0 | patient_id)
Data: dat
REML criterion at convergence: 888.5308
Random effects:
Groups Name Std.Dev. Corr
patient_id (Intercept) 0.0000
patient_id.1 timepointpre 0.3268
timepointpost 4.2039 -1.00
Residual 1.9086
Number of obs: 200, groups: patient_id, 20
Fixed Effects:
(Intercept) timepointpost
0.1868 1.0063
optimizer (nloptwrap) convergence code: 0 (OK) ; 0 optimizer warnings; 1 lme4 warnings